Use Seq2Seq Model to Predict the Calibrations¶
Settings:¶
Input: the previous calibrations and luminosity;
Output: the future calibrations;
Training data: the data collected from the crystal 54,000 in the year of 2016.
Two Cases:¶
Case1: We do not reuse the predicted values. For example, we do not use the predicted values at t to predict the t+1 values.
Case2: We do reuse the predicted values. For example, we do use the predicted values at t to predict the t+1 values.
Main:¶
This is the main function/entry for our Seq2Seq model.
Import packages/classes.¶
seq2seq_model defines the model;
seq2seq_train defines the training procedures;
seq2seq_prediction defines the prediction procedures;
ecal_dataset_prep defines the data preprocessing procedures.
[1]:
#------ import packages ------#
from seq2seq_model import *
from seq2seq_train import *
from seq2seq_prediction import *
from ecal_dataset_prep import *
Set up the hyper-parameters.¶
input_len: the time steps (sequence length) for input data;
output_len: the time steps (sequence length) for output data;
stride: the stide of the sequence/window (default: output_len);
learning_rate: the learning rate for our model;
n_epochs: the maximum epoch to train our model;
print_step: we print the training information per “print_step” epoch;
batch_size: the batch size to train our model;
opt_alg: the name of the optimization function (one should select one from {adam’, ‘sgd’});
train_strategy: different training strategies (one should select one from {‘recursive’, ‘teacher_forcing’, ‘mixed’});
teacher_forcing_ratio: it is a float number in the range of 0-1; it will be ignored when train_strategy=‘recursive’;
hidden_size: the number of features in the hidden state;
num_layers: the number of recurrent layers;
gpu_id: the gpu id;
train_file: the training csv file;
val_file: the validation csv file;
test_file: the test csv file;
crystal_id: the crystal’s id;
verbose:set it to be True if print information is desired; otherwise, set it to False; default (False).
[2]:
input_len = 24
output_len = 24
stride = output_len
learning_rate = 1e-3
n_epochs = 200
print_step = 1
batch_size = 128
if output_len>=48: batch_size = 32
opt_alg = 'adam'
train_strategy = 'recursive' #"Please select one of them---[recursive, teacher_forcing, mixed]!"
teacher_forcing_ratio = 0.5 # please set it in the range of [0,1]
hidden_size = 1024
num_layers = 2
gpu_id = 0
crystal_id = 54000
verbose = False
train_file_2016 = '../0_Dataset/interim/df_skimmed_xtal_{}_2016.csv'.format(crystal_id)
test_file_2017 = '../0_Dataset/interim/df_skimmed_xtal_{}_2017.csv'.format(crystal_id)
test_file_2018 = '../0_Dataset/interim/df_skimmed_xtal_{}_2018.csv'.format(crystal_id)
device = torch.device("cuda:{}".format(gpu_id) if torch.cuda.is_available() else "cpu")
Create folders to keep the results¶
[3]:
# folder to save figures
save_dir_vis_data = 'LSTM_{}_IW_{}_OW_{}_LR_{}_ID_{}/vis_data/'.format(hidden_size, input_len, output_len, learning_rate,crystal_id)
# folder to save models
save_dir_models = 'LSTM_{}_IW_{}_OW_{}_LR_{}_ID_{}/models/'.format(hidden_size, input_len, output_len, learning_rate,crystal_id)
# folders for case1
save_dir_case1_fig= 'LSTM_{}_IW_{}_OW_{}_LR_{}_ID_{}/case1_fig/'.format(hidden_size, input_len, output_len, learning_rate,crystal_id)
save_dir_case1_csv= 'LSTM_{}_IW_{}_OW_{}_LR_{}_ID_{}/case1_csv/'.format(hidden_size, input_len, output_len, learning_rate,crystal_id)
# folders for case2
save_dir_case2_fig= 'LSTM_{}_IW_{}_OW_{}_LR_{}_ID_{}/case2_fig/'.format(hidden_size, input_len, output_len, learning_rate,crystal_id)
save_dir_case2_csv= 'LSTM_{}_IW_{}_OW_{}_LR_{}_ID_{}/case2_csv/'.format(hidden_size, input_len, output_len, learning_rate,crystal_id)
dir_list = [save_dir_vis_data, save_dir_case1_fig, save_dir_case1_csv, save_dir_case2_fig, save_dir_case2_csv,save_dir_models]
for cur_dir in dir_list:
if not os.path.exists(cur_dir):
os.makedirs(cur_dir)
print('>>> {} has been created successfully!'.format(cur_dir))
else:
print('>>> {} is exist!'.format(cur_dir))
>>> LSTM_1024_IW_24_OW_24_LR_0.001_ID_54000/vis_data/ is exist!
>>> LSTM_1024_IW_24_OW_24_LR_0.001_ID_54000/case1_fig/ is exist!
>>> LSTM_1024_IW_24_OW_24_LR_0.001_ID_54000/case1_csv/ is exist!
>>> LSTM_1024_IW_24_OW_24_LR_0.001_ID_54000/case2_fig/ is exist!
>>> LSTM_1024_IW_24_OW_24_LR_0.001_ID_54000/case2_csv/ is exist!
>>> LSTM_1024_IW_24_OW_24_LR_0.001_ID_54000/models/ is exist!
Visualize the datasets (only show the calibration curve).¶
[4]:
# for train_file_2016
fig_name_cali = os.path.join(save_dir_vis_data, '2016_cali_original.png')
fig_name_scaled_cali = os.path.join(save_dir_vis_data, '2016_cali_scaled.png')
ecal_dataset_prep_train_2016 = ECAL_Dataset_Prep(train_file_2016,
input_len,
output_len,
stride,
fig_name_cali,
fig_name_scaled_cali,
verbose)
ecal_dataset_prep_train_2016.start_processing()
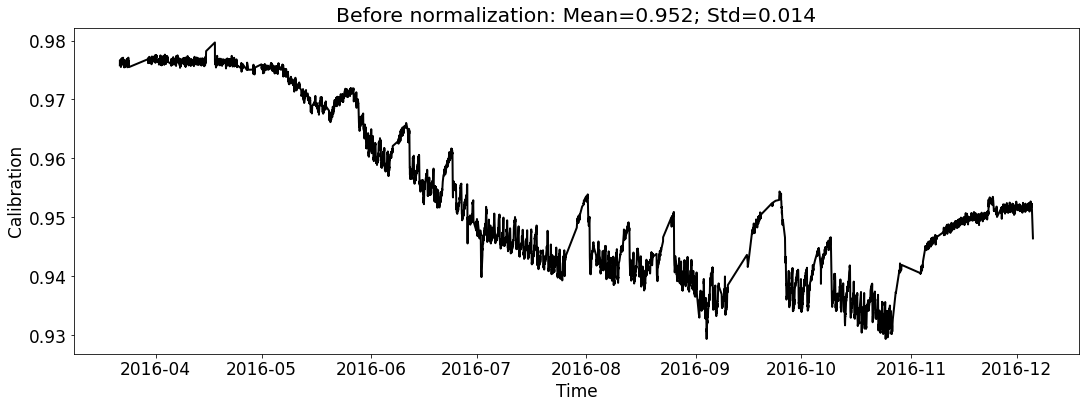
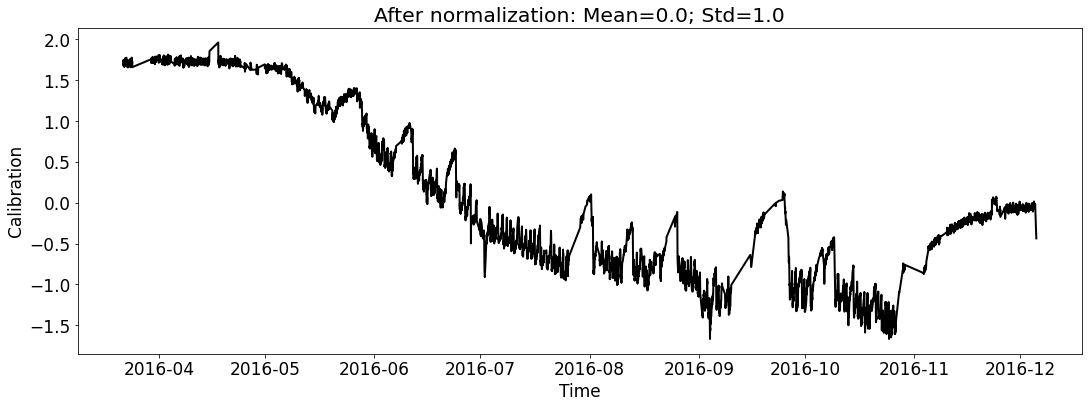
[5]:
# for test_file_2017
fig_name_cali = os.path.join(save_dir_vis_data, '2017_cali_original.png')
fig_name_scaled_cali = os.path.join(save_dir_vis_data, '2017_cali_scaled.png')
ecal_dataset_prep_test_2017 = ECAL_Dataset_Prep(test_file_2017,
input_len,
output_len,
stride,
fig_name_cali,
fig_name_scaled_cali,
verbose)
ecal_dataset_prep_test_2017.start_processing()
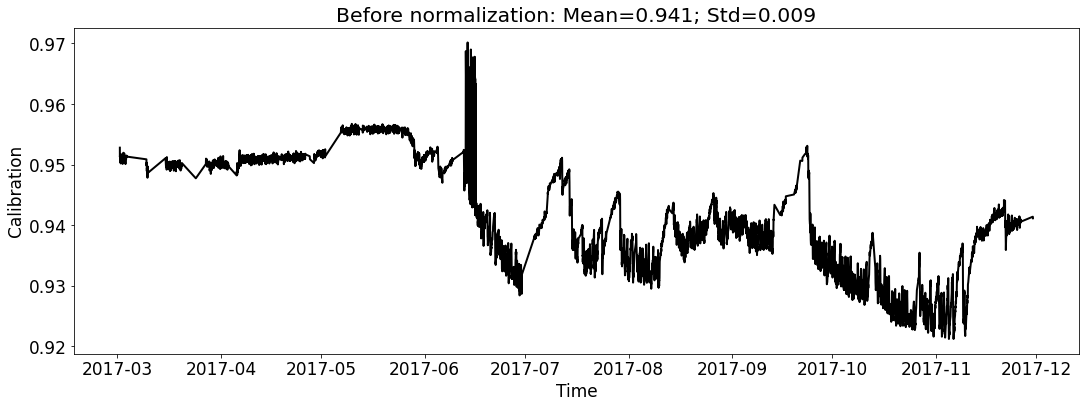
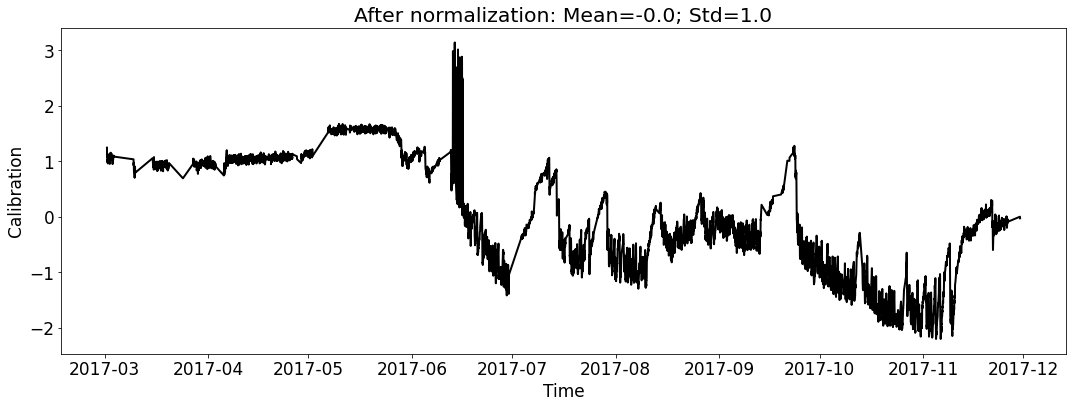
[6]:
# for test_file_2018
fig_name_cali = os.path.join(save_dir_vis_data, '2018_cali_original.png')
fig_name_scaled_cali = os.path.join(save_dir_vis_data, '2018_cali_scaled.png')
ecal_dataset_prep_test_2018 = ECAL_Dataset_Prep(test_file_2018,
input_len,
output_len,
stride,
fig_name_cali,
fig_name_scaled_cali,
verbose)
ecal_dataset_prep_test_2018.start_processing()
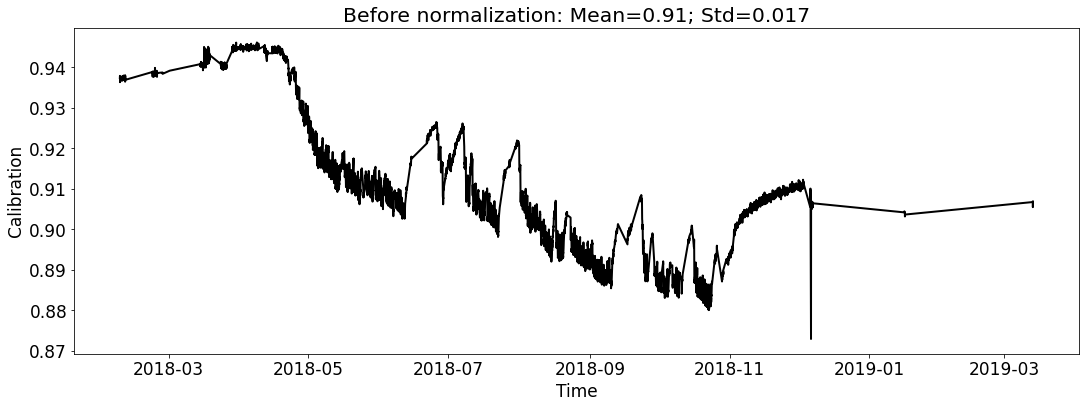
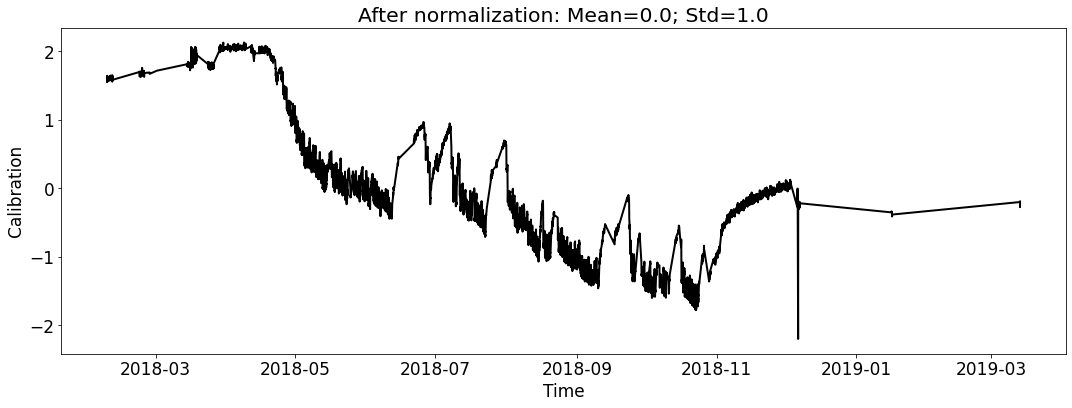
Create Seq2Seq model including an EncoderNet and a DecoderNet¶
[7]:
X_train = ecal_dataset_prep_train_2016.torch_X
Y_train = ecal_dataset_prep_train_2016.torch_Y
lstm_encoder = LSTM_Encoder(input_size=X_train.shape[2], hidden_size=hidden_size, num_layers=num_layers)
lstm_decoder = LSTM_Decoder(input_size=Y_train.shape[2], hidden_size=hidden_size, num_layers=num_layers)
lstm_encoder.to(device)
lstm_decoder.to(device)
print(lstm_encoder)
print(lstm_decoder)
LSTM_Encoder(
(lstm): LSTM(2, 1024, num_layers=2)
)
LSTM_Decoder(
(lstm): LSTM(2, 1024, num_layers=2)
(linear): Linear(in_features=1024, out_features=1, bias=True)
)
Training Seq2Seq model¶
[8]:
loss_figure_name = os.path.join(save_dir_vis_data, '0_loss.png')
target_len = output_len
seq2sqe_train = Seq2Seq_Train(lstm_encoder,
lstm_decoder,
X_train,
Y_train,
n_epochs,
target_len,
batch_size,
learning_rate,
opt_alg,
print_step,
train_strategy,
teacher_forcing_ratio,
device,
loss_figure_name)
seq2sqe_train.start_train()
>>> Start training... (be patient: training time varies)
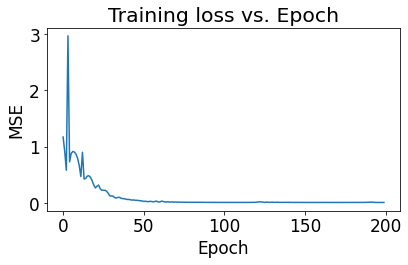
>>> Finish training!
Save trained models¶
[9]:
#after training, we also want to save our models
model_file_name = os.path.join(save_dir_models, 'lstm_encoder.pt')
save_model(lstm_encoder.eval(), model_file_name)
model_file_name = os.path.join(save_dir_models, 'lstm_decoder.pt')
save_model(lstm_decoder.eval(), model_file_name)
The trained model has been saved!
The trained model has been saved!
Case1 Prediction: do not use predictions as input to help the next-round prediction¶
[10]:
# check its prediction on training data
# Please note that here, the data are in the numpy format, not the tensor format
Xtrain = ecal_dataset_prep_train_2016.np_X
Ytrain = ecal_dataset_prep_train_2016.np_Y
df = ecal_dataset_prep_train_2016.df_lumi
scaler_cali = ecal_dataset_prep_train_2016.scaler_cali
year = '2016'
test_case = 'case1'
fig_name_mape = os.path.join(save_dir_case1_fig, '0_MAPE_{}_{}.png'.format(test_case,year))
fig_name_mse = os.path.join(save_dir_case1_fig, '1_MSE_{}_{}.png'.format(test_case,year))
metric_file = os.path.join(save_dir_case1_csv, '{}_{}.csv'.format(test_case,year))
seq2seq_prediction = Seq2Seq_Prediction(lstm_encoder,
lstm_decoder,
Xtrain,
Ytrain,
df,
scaler_cali,
device,
fig_name_mape,
fig_name_mse,
metric_file,
test_case)
seq2seq_prediction.start_prediction()
>>> case1 : start prediction...(be patient)
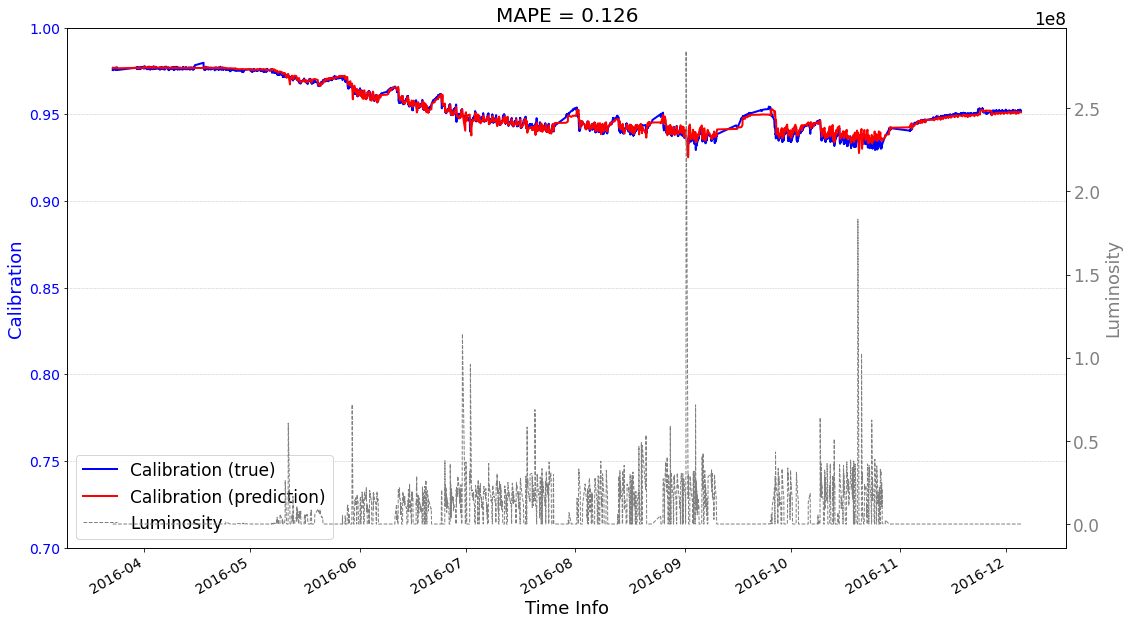
>>> Finish prediction!
[11]:
# check its prediction on test data-2017
# Please note that here, the data are in the numpy format, not the tensor format
Xtrain = ecal_dataset_prep_test_2017.np_X
Ytrain = ecal_dataset_prep_test_2017.np_Y
df = ecal_dataset_prep_test_2017.df_lumi
scaler_cali = ecal_dataset_prep_test_2017.scaler_cali
year = '2017'
test_case = 'case1'
fig_name_mape = os.path.join(save_dir_case1_fig, '0_MAPE_{}_{}.png'.format(test_case,year))
fig_name_mse = os.path.join(save_dir_case1_fig, '1_MSE_{}_{}.png'.format(test_case,year))
metric_file = os.path.join(save_dir_case1_csv, '{}_{}.csv'.format(test_case,year))
seq2seq_prediction = Seq2Seq_Prediction(lstm_encoder,
lstm_decoder,
Xtrain,
Ytrain,
df,
scaler_cali,
device,
fig_name_mape,
fig_name_mse,
metric_file,
test_case)
seq2seq_prediction.start_prediction()
>>> case1 : start prediction...(be patient)
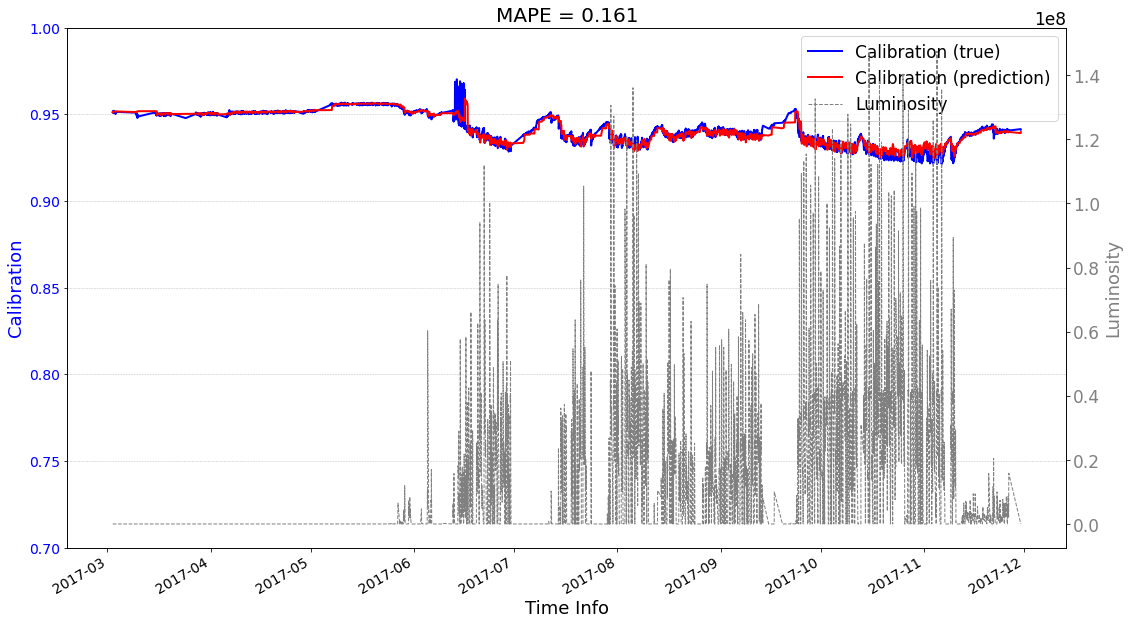
>>> Finish prediction!
[12]:
# check its prediction on test data-2018
# Please note that here, the data are in the numpy format, not the tensor format
Xtrain = ecal_dataset_prep_test_2018.np_X
Ytrain = ecal_dataset_prep_test_2018.np_Y
df = ecal_dataset_prep_test_2018.df_lumi
scaler_cali = ecal_dataset_prep_test_2017.scaler_cali
year = '2018'
test_case = 'case1'
fig_name_mape = os.path.join(save_dir_case1_fig, '0_MAPE_{}_{}.png'.format(test_case,year))
fig_name_mse = os.path.join(save_dir_case1_fig, '1_MSE_{}_{}.png'.format(test_case,year))
metric_file = os.path.join(save_dir_case1_csv, '{}_{}.csv'.format(test_case,year))
seq2seq_prediction = Seq2Seq_Prediction(lstm_encoder,
lstm_decoder,
Xtrain,
Ytrain,
df,
scaler_cali,
device,
fig_name_mape,
fig_name_mse,
metric_file,
test_case)
seq2seq_prediction.start_prediction()
>>> case1 : start prediction...(be patient)
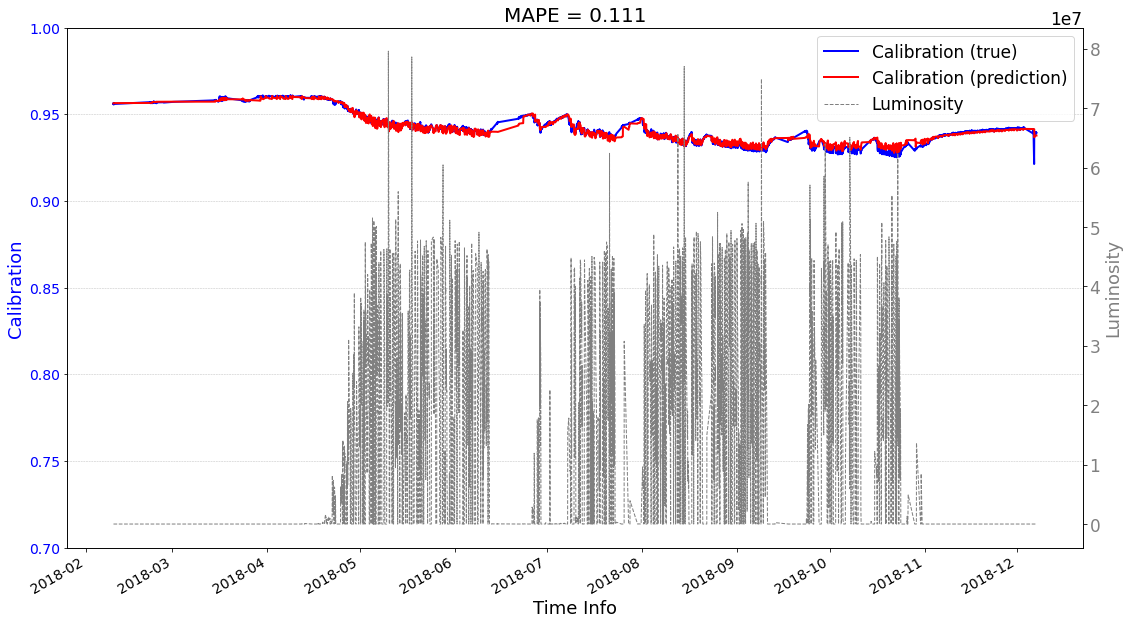
>>> Finish prediction!
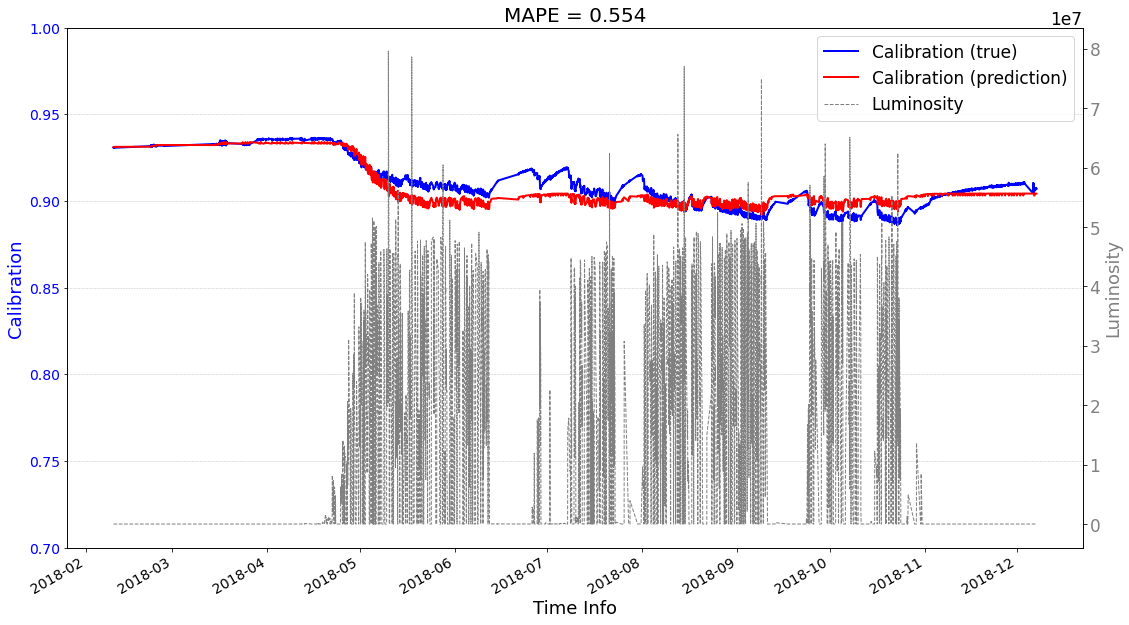
Case2 Prediction: use predictions as input to help the next-round prediction.¶
[13]:
# check its prediction on training data
# Please note that here, the data are in the numpy format, not the tensor format
Xtrain = ecal_dataset_prep_train_2016.np_X
Ytrain = ecal_dataset_prep_train_2016.np_Y
df = ecal_dataset_prep_train_2016.df_lumi
scaler_cali = ecal_dataset_prep_train_2016.scaler_cali
year = '2016'
test_case = 'case2'
fig_name_mape = os.path.join(save_dir_case1_fig, '0_MAPE_{}_{}.png'.format(test_case,year))
fig_name_mse = os.path.join(save_dir_case1_fig, '1_MSE_{}_{}.png'.format(test_case,year))
metric_file = os.path.join(save_dir_case1_csv, '{}_{}.csv'.format(test_case,year))
seq2seq_prediction = Seq2Seq_Prediction(lstm_encoder,
lstm_decoder,
Xtrain,
Ytrain,
df,
scaler_cali,
device,
fig_name_mape,
fig_name_mse,
metric_file,
test_case)
seq2seq_prediction.start_prediction()
>>> case2 : start prediction...(be patient)
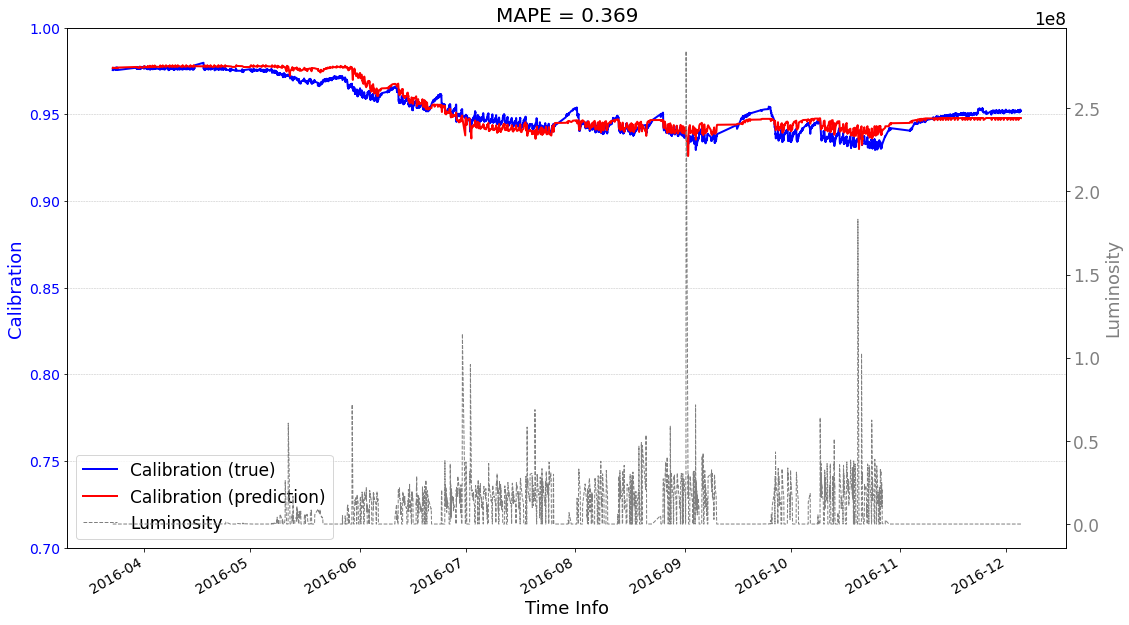
>>> Finish prediction!
[14]:
# check its prediction on test data-2017
# Please note that here, the data are in the numpy format, not the tensor format
Xtrain = ecal_dataset_prep_test_2017.np_X
Ytrain = ecal_dataset_prep_test_2017.np_Y
df = ecal_dataset_prep_test_2017.df_lumi
scaler_cali = ecal_dataset_prep_test_2017.scaler_cali
year = '2017'
test_case = 'case2'
fig_name_mape = os.path.join(save_dir_case1_fig, '0_MAPE_{}_{}.png'.format(test_case,year))
fig_name_mse = os.path.join(save_dir_case1_fig, '1_MSE_{}_{}.png'.format(test_case,year))
metric_file = os.path.join(save_dir_case1_csv, '{}_{}.csv'.format(test_case,year))
seq2seq_prediction = Seq2Seq_Prediction(lstm_encoder,
lstm_decoder,
Xtrain,
Ytrain,
df,
scaler_cali,
device,
fig_name_mape,
fig_name_mse,
metric_file,
test_case)
seq2seq_prediction.start_prediction()
>>> case2 : start prediction...(be patient)
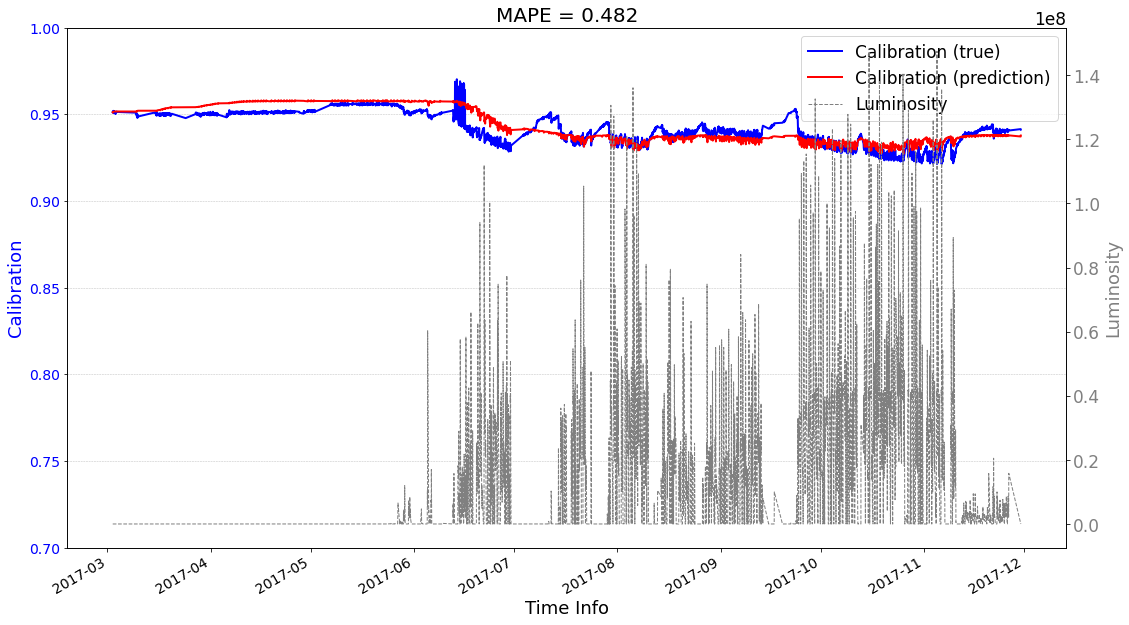
>>> Finish prediction!
[15]:
# check its prediction on test data-2018
# Please note that here, the data are in the numpy format, not the tensor format
Xtrain = ecal_dataset_prep_test_2018.np_X
Ytrain = ecal_dataset_prep_test_2018.np_Y
df = ecal_dataset_prep_test_2018.df_lumi
scaler_cali = ecal_dataset_prep_test_2017.scaler_cali
year = '2018'
test_case = 'case2'
fig_name_mape = os.path.join(save_dir_case1_fig, '0_MAPE_{}_{}.png'.format(test_case,year))
fig_name_mse = os.path.join(save_dir_case1_fig, '1_MSE_{}_{}.png'.format(test_case,year))
metric_file = os.path.join(save_dir_case1_csv, '{}_{}.csv'.format(test_case,year))
seq2seq_prediction = Seq2Seq_Prediction(lstm_encoder,
lstm_decoder,
Xtrain,
Ytrain,
df,
scaler_cali,
device,
fig_name_mape,
fig_name_mse,
metric_file,
test_case)
seq2seq_prediction.start_prediction()
>>> case2 : start prediction...(be patient)
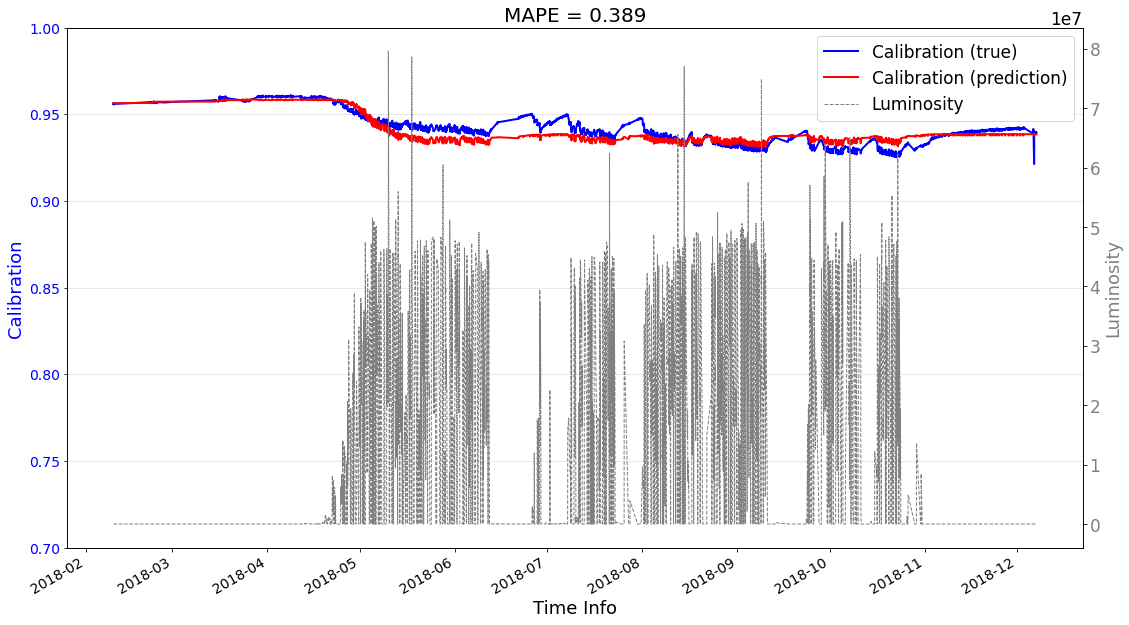
>>> Finish prediction!
Test on other crystals¶
[16]:
crystal_id = 54001
test_file_2016 = '../0_Dataset/interim/df_skimmed_xtal_{}_2016.csv'.format(crystal_id)
test_file_2017 = '../0_Dataset/interim/df_skimmed_xtal_{}_2017.csv'.format(crystal_id)
test_file_2018 = '../0_Dataset/interim/df_skimmed_xtal_{}_2018.csv'.format(crystal_id)
[17]:
# folders for case1
save_dir_case1_fig= 'LSTM_{}_IW_{}_OW_{}_LR_{}_ID_{}/case1_fig/'.format(hidden_size, input_len, output_len, learning_rate,crystal_id)
save_dir_case1_csv= 'LSTM_{}_IW_{}_OW_{}_LR_{}_ID_{}/case1_csv/'.format(hidden_size, input_len, output_len, learning_rate,crystal_id)
# folders for case2
save_dir_case2_fig= 'LSTM_{}_IW_{}_OW_{}_LR_{}_ID_{}/case2_fig/'.format(hidden_size, input_len, output_len, learning_rate,crystal_id)
save_dir_case2_csv= 'LSTM_{}_IW_{}_OW_{}_LR_{}_ID_{}/case2_csv/'.format(hidden_size, input_len, output_len, learning_rate,crystal_id)
dir_list = [save_dir_vis_data, save_dir_case1_fig, save_dir_case1_csv, save_dir_case2_fig, save_dir_case2_csv,save_dir_models]
for cur_dir in dir_list:
if not os.path.exists(cur_dir):
os.makedirs(cur_dir)
print('>>> {} has been created successfully!'.format(cur_dir))
else:
print('>>> {} is exist!'.format(cur_dir))
>>> LSTM_1024_IW_24_OW_24_LR_0.001_ID_54000/vis_data/ is exist!
>>> LSTM_1024_IW_24_OW_24_LR_0.001_ID_54001/case1_fig/ is exist!
>>> LSTM_1024_IW_24_OW_24_LR_0.001_ID_54001/case1_csv/ is exist!
>>> LSTM_1024_IW_24_OW_24_LR_0.001_ID_54001/case2_fig/ is exist!
>>> LSTM_1024_IW_24_OW_24_LR_0.001_ID_54001/case2_csv/ is exist!
>>> LSTM_1024_IW_24_OW_24_LR_0.001_ID_54000/models/ is exist!
[18]:
# for test_file_2016
fig_name_cali = os.path.join(save_dir_vis_data, '2016_cali_original.png')
fig_name_scaled_cali = os.path.join(save_dir_vis_data, '2016_cali_scaled.png')
ecal_dataset_prep_test_2016 = ECAL_Dataset_Prep(test_file_2016,
input_len,
output_len,
stride,
fig_name_cali,
fig_name_scaled_cali,
verbose)
ecal_dataset_prep_test_2016.start_processing()
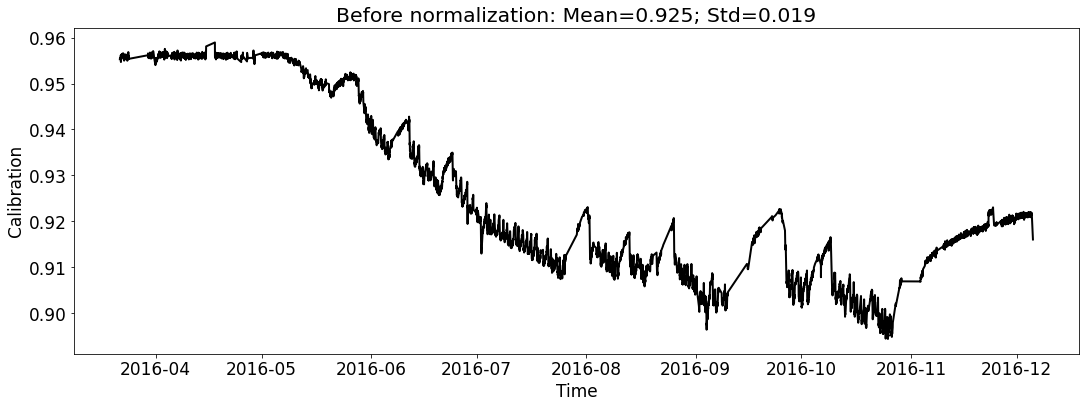
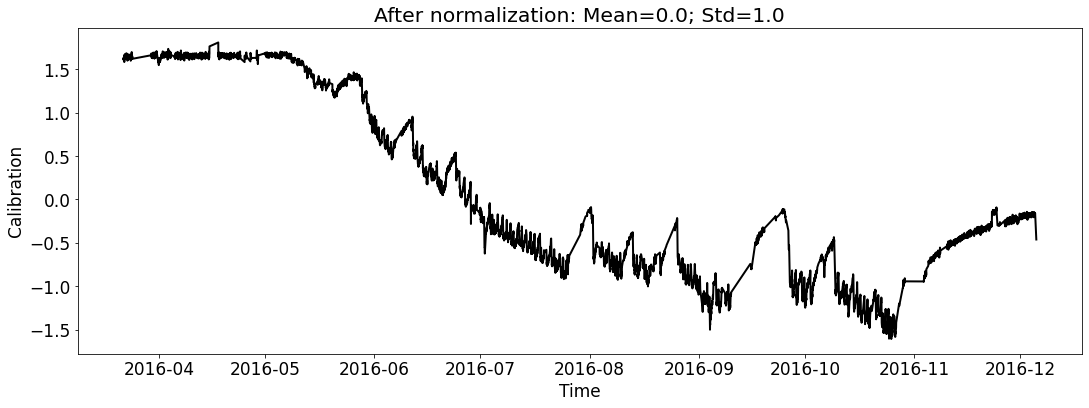
[19]:
# for test_file_2017
fig_name_cali = os.path.join(save_dir_vis_data, '2017_cali_original.png')
fig_name_scaled_cali = os.path.join(save_dir_vis_data, '2017_cali_scaled.png')
ecal_dataset_prep_test_2017 = ECAL_Dataset_Prep(test_file_2017,
input_len,
output_len,
stride,
fig_name_cali,
fig_name_scaled_cali,
verbose)
ecal_dataset_prep_test_2017.start_processing()
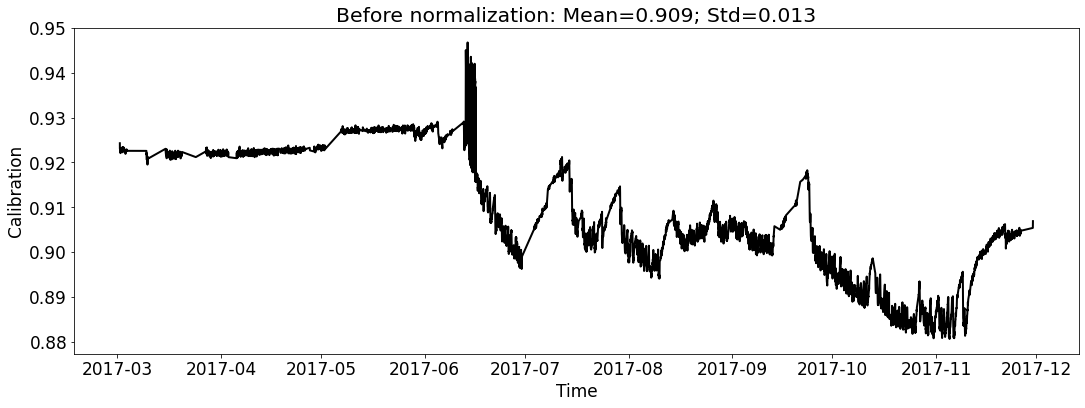
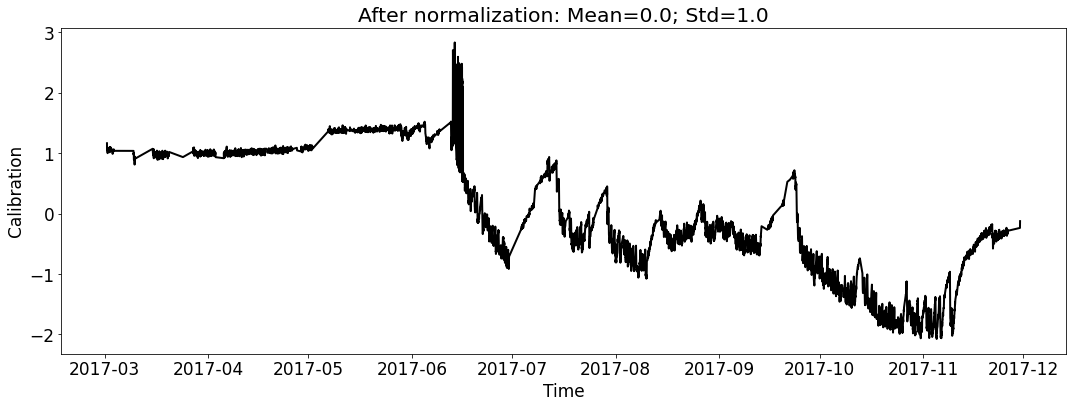
[20]:
# for test_file_2018
fig_name_cali = os.path.join(save_dir_vis_data, '2018_cali_original.png')
fig_name_scaled_cali = os.path.join(save_dir_vis_data, '2018_cali_scaled.png')
ecal_dataset_prep_test_2018 = ECAL_Dataset_Prep(test_file_2018,
input_len,
output_len,
stride,
fig_name_cali,
fig_name_scaled_cali,
verbose)
ecal_dataset_prep_test_2018.start_processing()
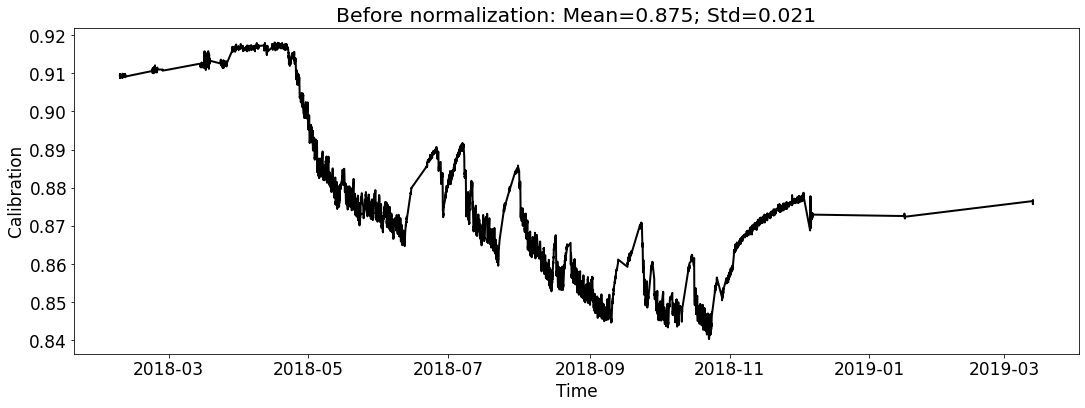
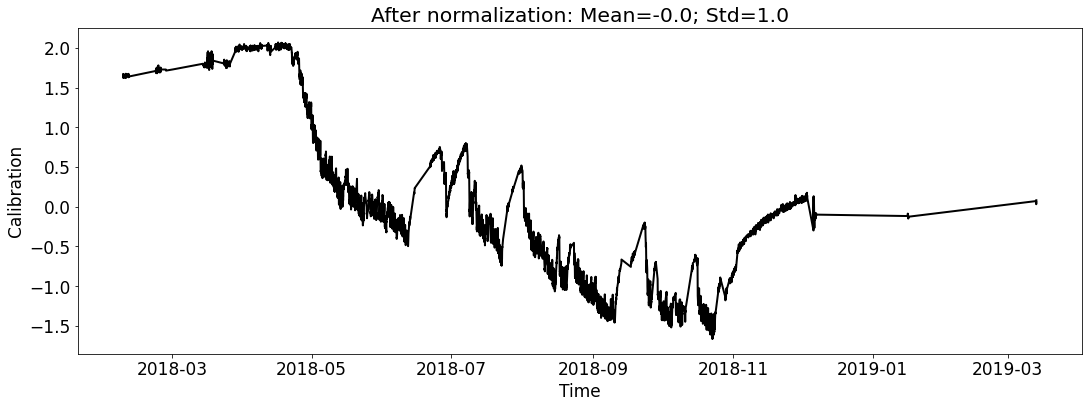
Case1 Prediction: do not use prediction as input to help the next-round prediction¶
[21]:
# check its prediction on test data
# Please note that here, the data are in the numpy format, not the tensor format
Xtrain = ecal_dataset_prep_test_2016.np_X
Ytrain = ecal_dataset_prep_test_2016.np_Y
df = ecal_dataset_prep_test_2016.df_lumi
scaler_cali = ecal_dataset_prep_test_2016.scaler_cali
year = '2016'
test_case = 'case1'
fig_name_mape = os.path.join(save_dir_case1_fig, '0_MAPE_{}_{}.png'.format(test_case,year))
fig_name_mse = os.path.join(save_dir_case1_fig, '1_MSE_{}_{}.png'.format(test_case,year))
metric_file = os.path.join(save_dir_case1_csv, '{}_{}.csv'.format(test_case,year))
seq2seq_prediction = Seq2Seq_Prediction(lstm_encoder,
lstm_decoder,
Xtrain,
Ytrain,
df,
scaler_cali,
device,
fig_name_mape,
fig_name_mse,
metric_file,
test_case)
seq2seq_prediction.start_prediction()
>>> case1 : start prediction...(be patient)
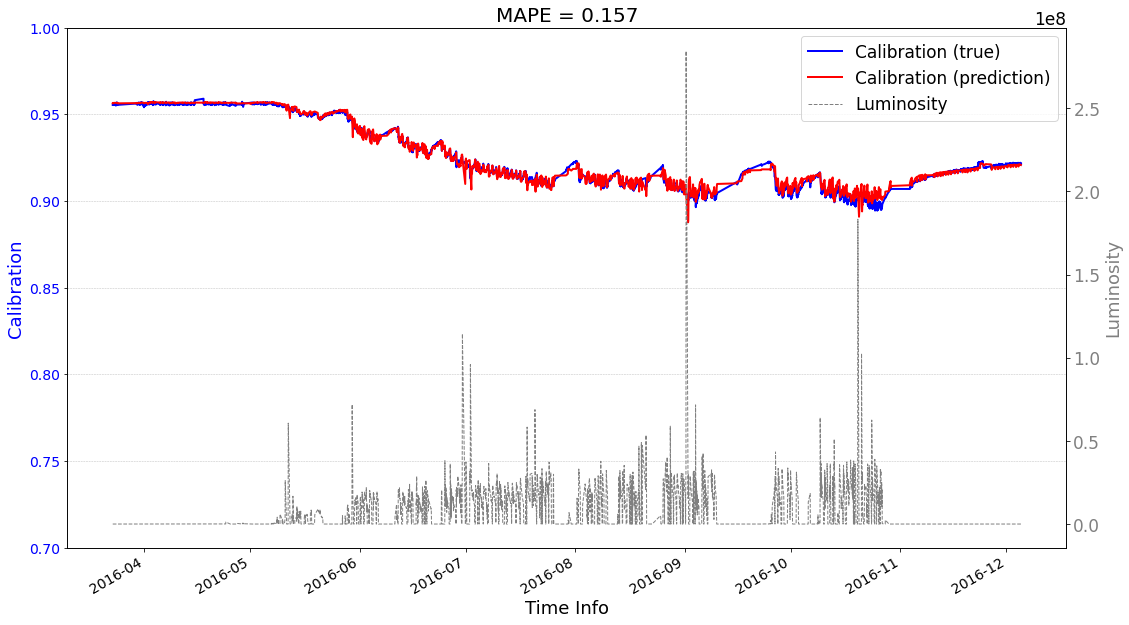
>>> Finish prediction!
[22]:
# check its prediction on test data-2017
# Please note that here, the data are in the numpy format, not the tensor format
Xtrain = ecal_dataset_prep_test_2017.np_X
Ytrain = ecal_dataset_prep_test_2017.np_Y
df = ecal_dataset_prep_test_2017.df_lumi
scaler_cali = ecal_dataset_prep_test_2017.scaler_cali
year = '2017'
test_case = 'case1'
fig_name_mape = os.path.join(save_dir_case1_fig, '0_MAPE_{}_{}.png'.format(test_case,year))
fig_name_mse = os.path.join(save_dir_case1_fig, '1_MSE_{}_{}.png'.format(test_case,year))
metric_file = os.path.join(save_dir_case1_csv, '{}_{}.csv'.format(test_case,year))
seq2seq_prediction = Seq2Seq_Prediction(lstm_encoder,
lstm_decoder,
Xtrain,
Ytrain,
df,
scaler_cali,
device,
fig_name_mape,
fig_name_mse,
metric_file,
test_case)
seq2seq_prediction.start_prediction()
>>> case1 : start prediction...(be patient)
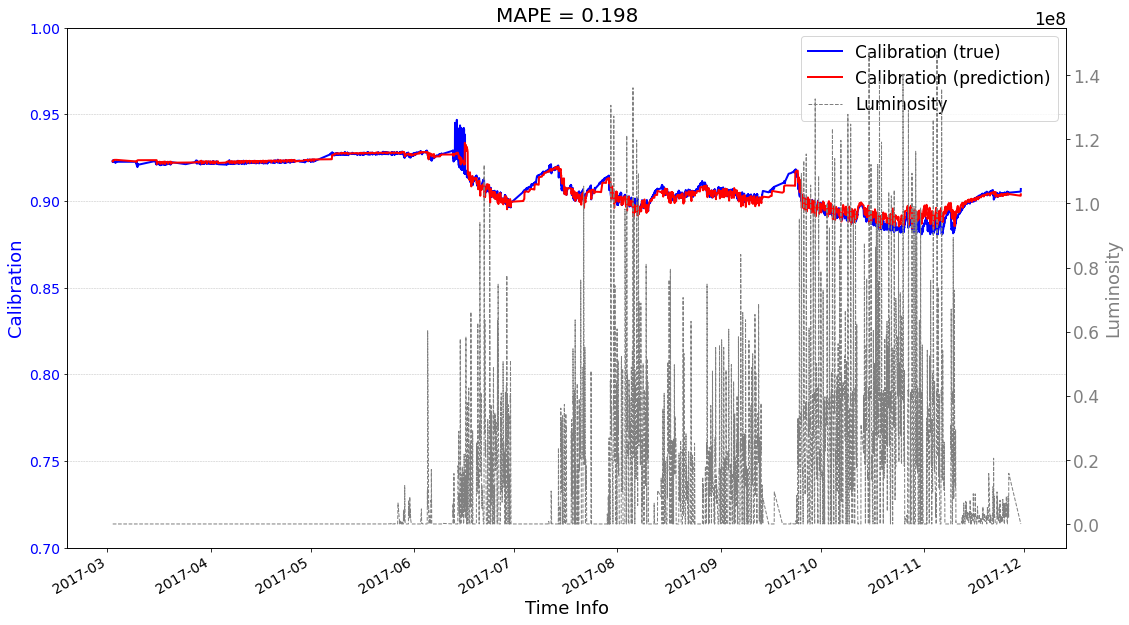
>>> Finish prediction!
[23]:
# check its prediction on test data-2018
# Please note that here, the data are in the numpy format, not the tensor format
Xtrain = ecal_dataset_prep_test_2018.np_X
Ytrain = ecal_dataset_prep_test_2018.np_Y
df = ecal_dataset_prep_test_2018.df_lumi
scaler_cali = ecal_dataset_prep_test_2017.scaler_cali
year = '2018'
test_case = 'case1'
fig_name_mape = os.path.join(save_dir_case1_fig, '0_MAPE_{}_{}.png'.format(test_case,year))
fig_name_mse = os.path.join(save_dir_case1_fig, '1_MSE_{}_{}.png'.format(test_case,year))
metric_file = os.path.join(save_dir_case1_csv, '{}_{}.csv'.format(test_case,year))
seq2seq_prediction = Seq2Seq_Prediction(lstm_encoder,
lstm_decoder,
Xtrain,
Ytrain,
df,
scaler_cali,
device,
fig_name_mape,
fig_name_mse,
metric_file,
test_case)
seq2seq_prediction.start_prediction()
>>> case1 : start prediction...(be patient)
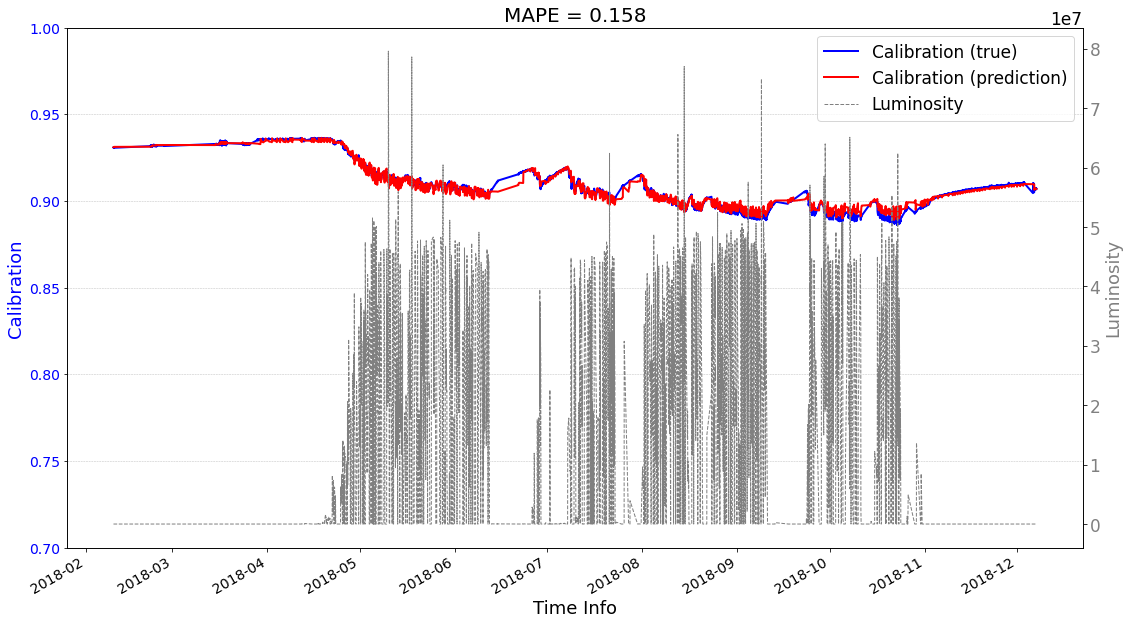
>>> Finish prediction!
Case2 Prediction: use predictions as input to help the next-round prediction.¶
[24]:
# check its prediction on test data
# Please note that here, the data are in the numpy format, not the tensor format
Xtrain = ecal_dataset_prep_test_2016.np_X
Ytrain = ecal_dataset_prep_test_2016.np_Y
df = ecal_dataset_prep_test_2016.df_lumi
scaler_cali = ecal_dataset_prep_test_2016.scaler_cali
year = '2016'
test_case = 'case2'
fig_name_mape = os.path.join(save_dir_case1_fig, '0_MAPE_{}_{}.png'.format(test_case,year))
fig_name_mse = os.path.join(save_dir_case1_fig, '1_MSE_{}_{}.png'.format(test_case,year))
metric_file = os.path.join(save_dir_case1_csv, '{}_{}.csv'.format(test_case,year))
seq2seq_prediction = Seq2Seq_Prediction(lstm_encoder,
lstm_decoder,
Xtrain,
Ytrain,
df,
scaler_cali,
device,
fig_name_mape,
fig_name_mse,
metric_file,
test_case)
seq2seq_prediction.start_prediction()
>>> case2 : start prediction...(be patient)
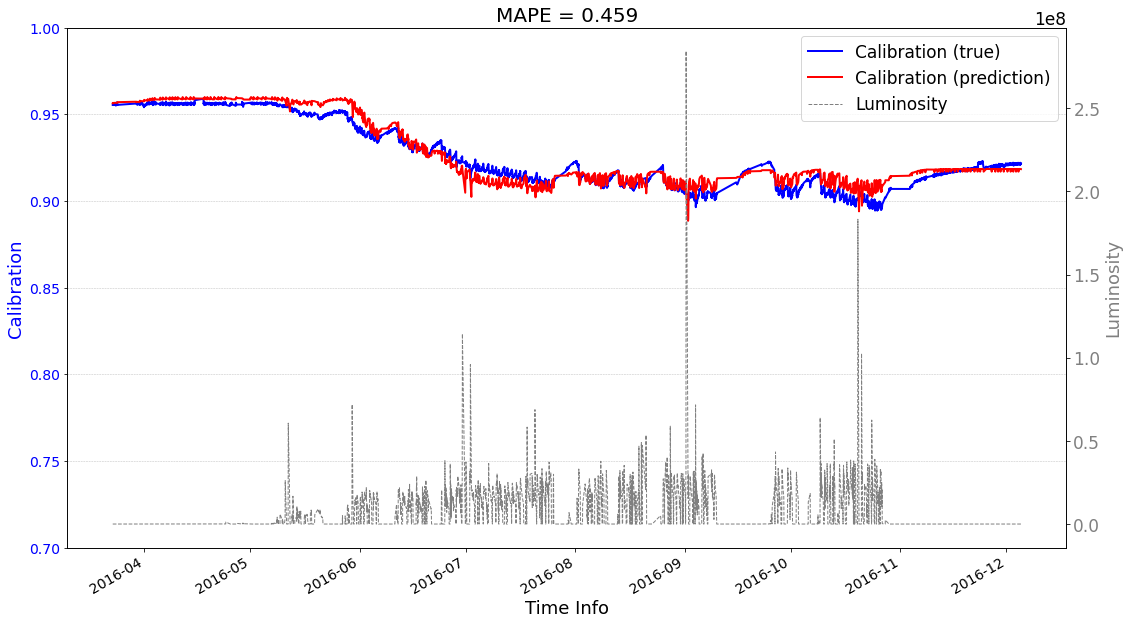
>>> Finish prediction!
[25]:
# check its prediction on test data-2017
# Please note that here, the data are in the numpy format, not the tensor format
Xtrain = ecal_dataset_prep_test_2017.np_X
Ytrain = ecal_dataset_prep_test_2017.np_Y
df = ecal_dataset_prep_test_2017.df_lumi
scaler_cali = ecal_dataset_prep_test_2017.scaler_cali
year = '2017'
test_case = 'case2'
fig_name_mape = os.path.join(save_dir_case1_fig, '0_MAPE_{}_{}.png'.format(test_case,year))
fig_name_mse = os.path.join(save_dir_case1_fig, '1_MSE_{}_{}.png'.format(test_case,year))
metric_file = os.path.join(save_dir_case1_csv, '{}_{}.csv'.format(test_case,year))
seq2seq_prediction = Seq2Seq_Prediction(lstm_encoder,
lstm_decoder,
Xtrain,
Ytrain,
df,
scaler_cali,
device,
fig_name_mape,
fig_name_mse,
metric_file,
test_case)
seq2seq_prediction.start_prediction()
>>> case2 : start prediction...(be patient)
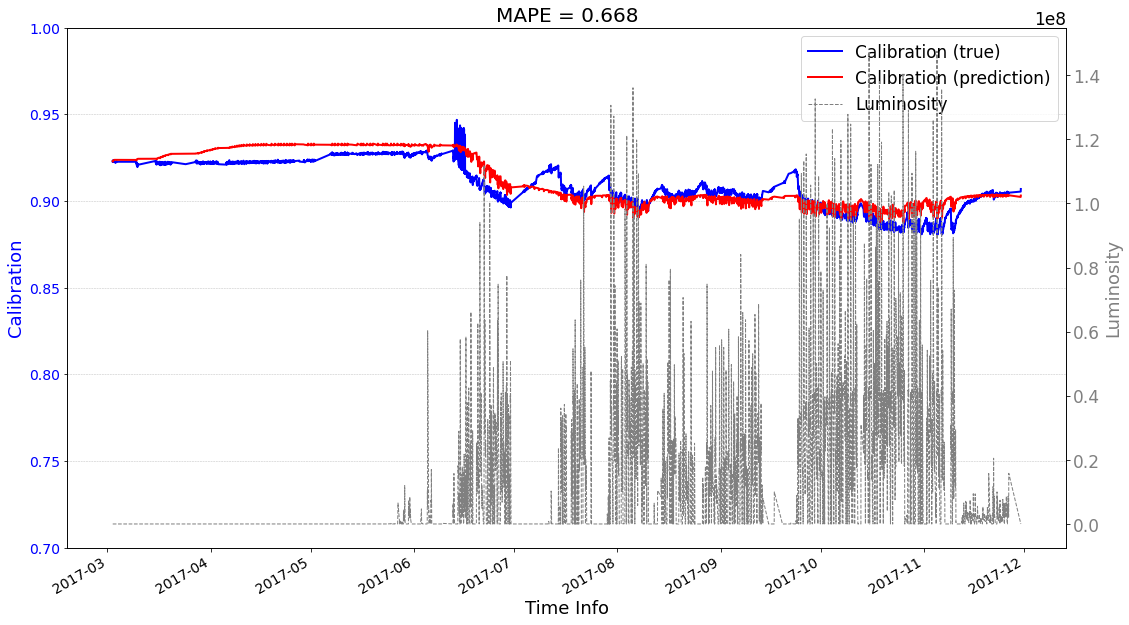
>>> Finish prediction!
[26]:
# check its prediction on test data-2018
# Please note that here, the data are in the numpy format, not the tensor format
Xtrain = ecal_dataset_prep_test_2018.np_X
Ytrain = ecal_dataset_prep_test_2018.np_Y
df = ecal_dataset_prep_test_2018.df_lumi
scaler_cali = ecal_dataset_prep_test_2017.scaler_cali
year = '2018'
test_case = 'case2'
fig_name_mape = os.path.join(save_dir_case1_fig, '0_MAPE_{}_{}.png'.format(test_case,year))
fig_name_mse = os.path.join(save_dir_case1_fig, '1_MSE_{}_{}.png'.format(test_case,year))
metric_file = os.path.join(save_dir_case1_csv, '{}_{}.csv'.format(test_case,year))
seq2seq_prediction = Seq2Seq_Prediction(lstm_encoder,
lstm_decoder,
Xtrain,
Ytrain,
df,
scaler_cali,
device,
fig_name_mape,
fig_name_mse,
metric_file,
test_case)
seq2seq_prediction.start_prediction()
>>> case2 : start prediction...(be patient)
>>> Finish prediction!
